研究揭示土壤中新型抗生素来源
来源:《自然》
作者:Alexander Crits-Christoph等
时间:2018-07-11
一项研究表明,土壤中的细菌可能代表了一种有待开发的新型抗生素和其他药用化合物的来源。该成果近日在线发表于 《自然》。
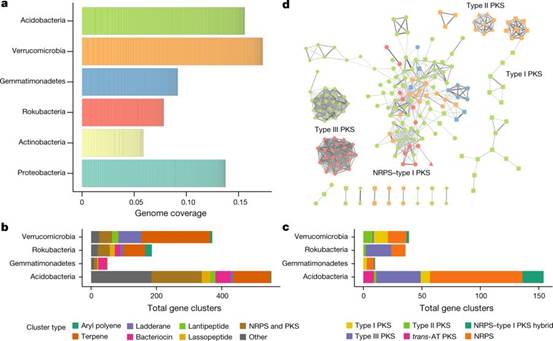
美国加州大学伯克利分校的 Jillian Banfield 及同事绘制了数百个非培养且基本未被研究的微生物基因组草图,这些微生物来自加利福尼亚州北部一片草原的土壤生态系统。他们鉴定出了 1000 多个生物合成基因簇,推测它们可合成一系列分子,包括各种肽、抗菌毒素和功能不明的其他代谢物。这些基因的表达因土壤样品采集的深度和位置不同而变化,反映了它们对不同环境条件的生物反应。
已知土壤中的微生物会产生多种有用的次级代谢物,包括抗生素、抗真菌剂和免疫抑制剂,但是它们大多数都来源于少数培养的微生物群。这项研究扩大了这些分子的潜在来源范围,甚至还发现了两个以前未知的具有强大生物合成能力的细菌种类。此外,这项研究也提出了土壤微生物使用这种复杂的化学语言相互沟通的可能性。(来源:中国科学报)
Novel soil bacteria possess diverse genes for secondary metabolite biosynthesis
Abstract In soil ecosystems, microorganisms produce diverse secondary metabolites such as antibiotics, antifungals and siderophores that mediate communication, competition and interactions with other organisms and the environment1,2. Most known antibiotics are derived from a few culturable microbial taxa3, and the biosynthetic potential of the vast majority of bacteria in soil has rarely been investigated4. Here we reconstruct hundreds of near-complete genomes from grassland soil metagenomes and identify microorganisms from previously understudied phyla that encode diverse polyketide and nonribosomal peptide biosynthetic gene clusters that are divergent from well-studied clusters. These biosynthetic loci are encoded by newly identified members of the Acidobacteria, Verrucomicobia and Gemmatimonadetes, and the candidate phylum Rokubacteria. Bacteria from these groups are highly abundant in soils5,6,7, but have not previously been genomically linked to secondary metabolite production with confidence. In particular, large numbers of biosynthetic genes were characterized in newly identified members of the Acidobacteria, which is the most abundant bacterial phylum across soil biomes5. We identify two acidobacterial genomes from divergent lineages, each of which encodes an unusually large repertoire of biosynthetic genes with up to fifteen large polyketide and nonribosomal peptide biosynthetic loci per genome. To track gene expression of genes encoding polyketide synthases and nonribosomal peptide synthetases in the soil ecosystem that we studied, we sampled 120 time points in a microcosm manipulation experiment and, using metatranscriptomics, found that gene clusters were differentially co-expressed in response to environmental perturbations. Transcriptional co-expression networks for specific organisms associated biosynthetic genes with two-component systems, transcriptional activation, putative antimicrobial resistance and iron regulation, linking metabolite biosynthesis to processes of environmental sensing and ecological competition. We conclude that the biosynthetic potential of abundant and phylogenetically diverse soil microorganisms has previously been underestimated. These organisms may represent a source of natural products that can address needs for new antibiotics and other pharmaceutical compounds.
原文链接:https://www.nature.com/articles/s41586-018-0207-y.pdf




