利用单细胞分析揭示蝾螈再生肢体机制
来源:《科学》
作者:Tobias Gerber等
时间:2018-11-19
美西螈(Ambystoma mexicanum, 也称钝口螈,六角恐龙)和其他的蝾螈是唯一能够再生全肢的四足动物。在这个复杂的过程中,基因表达的变化调节着新的附属肢体长出,但是人们对损伤如何诱导肢体细胞形成能够分化为多种细胞类型的再生性祖细胞知之甚少。在肢体再生过程中,对单个细胞进行追踪和分子分析将会鉴定出不同的分化途径,并为细胞如何从成熟的静息状态转变为再生性细胞谱系提供线索。
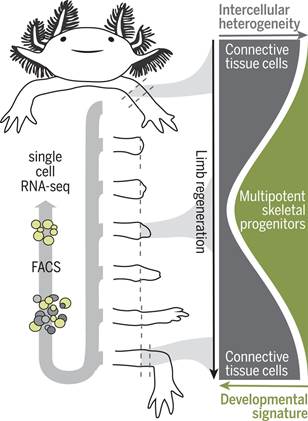
美西螈的肢体由许多不同的细胞类型组成,这些细胞类型源自神经细胞谱系、肌源性细胞谱系、表皮细胞谱系和结缔组织(connective tissue, CT)细胞谱系。肢体截肢后,来自截肢平面附近的细胞聚集在一个称为芽基(blastema)的独特组织中,芽基起着作为再生新肢体的祖细胞来源的作用。
在一种转基因美西螈品系中,不同的成体细胞类型的后代能够在再生过程中被标记、追踪和分离出来,这就为理解特定的细胞谱系在芽基形成和随后的肢体再生期间是如何发育的提供了机会。将转基因美西螈品系与单细胞RNA测序(scRNA-seq)相结合能够追踪单个细胞类型,以及重建这些特定的细胞谱系的再生过程背后的分子步骤。
作为侧板中胚层(lateral plate mesoderm)的后代,CT细胞是最为丰富的细胞谱系,有助于促进芽基产生,并且包围着骨骼和软骨、肌腱、骨骼外周(periskeleton)以及真皮成纤维细胞和间质成纤维细胞。这些细胞检测截肢部位所在的位置,导致适当的肢体部分再生,从而使得CT细胞成为破译和理解再生分子程序的关键细胞谱系。
在一项新的研究中,德国研究人员采用一种诱导型Cre-loxP荧光系统建立遗传标记的转基因美西螈品系用于分离成体肢体组织中的CT细胞和芽基中的CT细胞后代。他们利用scRNA-seq沿着芽基形成和再生胳膊长出的密集时间过程以及胚胎肢体的发育阶段对CT细胞进行分子分析。这种分子分析表明CT细胞表达一进入诱导再生时就失去的成体表型。这种源自CT细胞的异质细胞群体会聚到一种均匀而又短暂的芽基祖细胞状态,这种状态在后面的阶段能够重现胚胎肢体出芽样程序。相关研究结果发表在2018年10月26日的Science期刊上,论文标题为“Single-cell analysis uncovers convergence of cell identities during axolotl limb regeneration”。
值得注意的是,这些研究人员并没有在成熟的胳膊中发现CT干细胞或芽基样前体细胞存在的证据。他们发现CT细胞亚型对再生胳膊中的近端隔室和远端隔室作出空间限制的贡献。具体而言,一种特定的CT细胞亚型---骨骼外周细胞(periskeletal cell)---在截肢部位延伸切断的骨骼,然而成纤维细胞性的CT细胞从头再生远端的骨骼节段。
通过使用高通量单细胞转录组学分析和一种基于美西螈的脑彩虹克隆谱系追踪技术,这些研究人员能够在再生的最后阶段追踪CT细胞谱系的再分化轨迹。
这些发现确立了多能骨骼祖细胞(multipotent skeletal progenitor cell)的形成,而这些多能骨骼祖细胞导致肌腱、韧带、骨骼、骨骼外周和成纤维细胞产生。
综上所述,CT细胞是理解肢体再生的关键细胞类型,这是因为它们形成图案化的肢体骨骼,从而指导其他肢体组织(比如肌肉)的再生。鉴于CT细胞的异质性以及与其他细胞类型混合在一起,人们通常很难研究CT细胞如何形成再生性芽基细胞。对这些新培育出的转基因美西螈报告品系的使用,再结合单细胞转录组学分析和克隆追踪,允许这些研究人员确定出当具有不同分子特征的CT细胞去分化为一种常见的类似于胚胎肢芽细胞的多能祖细胞时,它们经历一种独特的分子状态。
在未来,测试这种过渡状态中的哪些组分是这种去分化过程所必需的将是比较重要的。此外,这项研究为探究再生相关基因及其相关的染色质结构在这种过渡过程中如何受到调节提供了可能性。最后,这项研究也提供了这样的一种可能性:在哺乳动物动物中观察到的有限再生能力是由于无法将CT细胞重编程到这样的胚胎状态。(来源:生物谷 Bioon.com)
Single-cell analysis uncovers convergence of cell identities during axolotl limb regeneration
Abstract INTRODUCTION Axolotls (Ambystoma mexicanum) and other salamanders are the only tetrapods that can regenerate whole limbs. During this complex process, changes in gene expression regulate the outgrowth of a new appendage, but how injury induces limb cells to form regenerative progenitors that differentiate into diverse cell fates is poorly understood. Tracking and molecular profiling of individual cells during limb regeneration would resolve distinct differentiation pathways and provide clues for how cells convert from a mature resting state into regenerative cell lineages. RATIONALE Axolotl limbs are composed of many different cell types originating from neural, myogenic, epidermal, and connective tissue (CT) lineages. Upon limb amputation, cells from nearby the amputation plane accumulate in a distinctive tissue called the blastema, which serves as a progenitor cell source to build the new limb. Transgenic axolotl strains in which descendants of distinct adult cell types can be labeled, tracked, and isolated during the regenerative process provide an opportunity to understand how particular cell lineages progress during blastema formation and subsequent limb regrowth. Combining transgenic axolotl strains with single-cell RNA sequencing (scRNA-seq) enables the tracking of individual cell types, as well as the reconstruction of the molecular steps underlying the regeneration process for these particular cell lineages. CT cells, descendants of lateral plate mesoderm, are the most abundant lineage contributing to the blastema and encompass bone and cartilage, tendons, periskeleton, and dermal and interstitial fibroblasts. These cells detect the position of the amputation site, leading to the regeneration of appropriate limb parts and making the CT a key cell lineage for deciphering and understanding molecular programs of regeneration. RESULTS We used an inducible Cre-loxP fluorescence system to establish genetically marked transgenic axolotl strains for isolating CT cells from adult limb tissue as well as CT descendants in the blastema. We used scRNA-seq to molecularly profile CT cells along a dense time course of blastema formation and the outgrowth of the regenerated arm, as well as stages of embryonic limb development. This profiling indicated that CT cells express adult phenotypes that are lost upon the induction of regeneration. The heterogeneous population of CT-derived cells converges into a homogeneous and transient blastema progenitor state that at later stages recapitulates an embryonic limb bud–like program. Notably, we did not find evidence of CT stem cells or blastema-like precursors in the mature arm. We found that CT subtypes have spatially restricted contributions to proximal and distal compartments in the regenerated limb. Specifically, a particular CT subtype—periskeletal cells—extended the severed skeleton at the amputation site whereas fibroblastic CT cells de novo regenerated distal skeletal segments. By using high-throughput single-cell transcriptomics and Brainbow axolotl-based clonal lineage tracing, we could follow the redifferentiation trajectories of CT lineages during the final stages of regeneration. These findings established the formation of a multipotent skeletal progenitor cell that contributed to tendons, ligaments, skeleton, periskeleton, and fibroblasts. CONCLUSION CT cells are a key cell type for understanding regeneration because they form the patterned limb skeleton that guides the regeneration of the other limb tissues, such as muscle. Because of the cells’ heterogeneity and intermingling with other cell types, it had been difficult to study how CT forms regenerative blastema cells. The use of these newly generated transgenic reporter strains combined with single-cell transcriptomic analysis and clonal tracing have allowed us to determine that CT cells with diverse molecular features traverse through a distinctive molecular state as they dedifferentiate into a common, multipotent progenitor resembling an embryonic limb bud cell. In the future, it will be important to test which components of the transition state are required for the dedifferentiation process. Furthermore, this work opens the possibility to examine how regeneration-associated genes and their associated chromatin structure are regulated during this transition. Lastly, the work raises the possibility that the limited regeneration seen among mammals is due to an inability to reprogram CT to such embryonic states.
原文链接:http://science.sciencemag.org/content/362/6413/eaaq0681/tab-pdf




