
英国《自然》杂志近日发表了两篇基因学论文,欧洲两组团队分别报告了美西螈和真涡虫的基因组,揭示了神秘“再生能力”背后的遗传学基础。其中美西螈的320亿个碱基对,是目前组装出的最大基因组。
美西螈全部肢体都可以再生,而真涡虫甚至可以在被切成碎块后,重新长出整个身体。研究人员一直都想彻底了解这其中的奥秘,弄清这种人类不具备的“再生能力”背后根本的遗传机制。
此次,奥地利分子病理学研究所的科学家团队测序了美西螈的基因组,它含有320亿个碱基对,是人类基因组的10倍,也是目前组装出的最大基因组。研究人员将那些在再生肢体细胞中表达较丰富的基因和miRNA序列标记为进一步研究的目标,并且发现,美西螈缺少Pax3基因——一种对许多其它动物的发育至关重要的基因。
德国马普学会分子细胞生物学和遗传学研究所的团队则测序了真涡虫的基因组,它含有约8亿碱基对。真涡虫的基因组中缺少大约124种对人类和小鼠至关重要的基因,包括参与DNA修复的基因以及帮助染色体在细胞分裂期间正确分离的基因。
美西螈和真涡虫的基因组都包含高水平的重复DNA序列,因而很难进行对其分析。科学家采用了一种新的计算方法,并结合长读长测序手段,改进了基因组组装。现在已知其某些重复序列参与到了胚胎发育和干细胞活动当中,它们是否就是美西螈和真涡虫身上神奇能力的关键,正是研究人员下一步研究的重点。(来源:科技日报 张梦然)
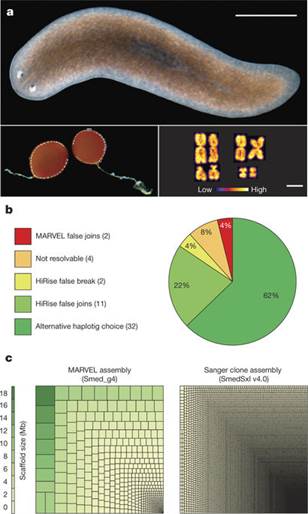
The genome of Schmidtea mediterranea and the evolution of core cellular mechanisms
Abstract The planarian Schmidtea mediterranea is an important model for stem cell research and regeneration, but adequate genome resources for this species have been lacking. Here we report a highly contiguous genome assembly of S. mediterranea, using long-read sequencing and a de novo assembler (MARVEL) enhanced for low-complexity reads. The S. mediterranea genome is highly polymorphic and repetitive, and harbours a novel class of giant retroelements. Furthermore, the genome assembly lacks a number of highly conserved genes, including critical components of the mitotic spindle assembly checkpoint, but planarians maintain checkpoint function. Our genome assembly provides a key model system resource that will be useful for studying regeneration and the evolutionary plasticity of core cell biological mechanisms.
原文链接:https://www.nature.com/articles/nature25473
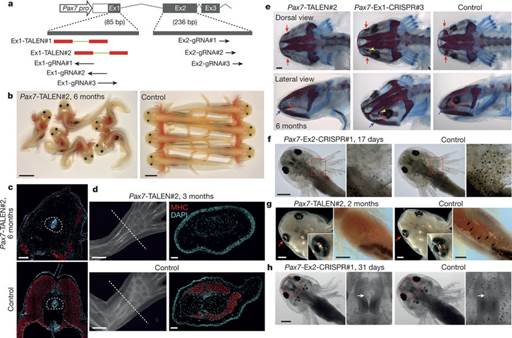
The axolotl genome and the evolution of key tissue formation regulators
Abstract Salamanders serve as important tetrapod models for developmental, regeneration and evolutionary studies. An extensive molecular toolkit makes the Mexican axolotl (Ambystoma mexicanum) a key representative salamander for molecular investigations. Here we report the sequencing and assembly of the 32-gigabase-pair axolotl genome using an approach that combined long-read sequencing, optical mapping and development of a new genome assembler (MARVEL). We observed a size expansion of introns and intergenic regions, largely attributable to multiplication of long terminal repeat retroelements. We provide evidence that intron size in developmental genes is under constraint and that species-restricted genes may contribute to limb regeneration. The axolotl genome assembly does not contain the essential developmental gene Pax3. However, mutation of the axolotl Pax3 paralogue Pax7 resulted in an axolotl phenotype that was similar to those seen in Pax3−/− and Pax7−/− mutant mice. The axolotl genome provides a rich biological resource for developmental and evolutionary studies.
原文链接:https://www.nature.com/articles/nature25458#auth-18



